maint_debian-med package set for trixie/arm64
Debian package sets:
desktop package sets:
Debian distribution package sets:
maintenance team package sets:
-
 maint_debian-accessibility
maint_debian-accessibility
-
 maint_debian-boot
maint_debian-boot
-
 maint_debian-lua
maint_debian-lua
-
 maint_debian-med
maint_debian-med
-
 maint_debian-ocaml
maint_debian-ocaml
-
 maint_debian-on-mobile-maintainers
maint_debian-on-mobile-maintainers
-
 maint_debian-python
maint_debian-python
-
 maint_debian-qa
maint_debian-qa
-
 maint_debian-science
maint_debian-science
-
 maint_debian-x
maint_debian-x
-
 maint_pkg-android-tools-devel
maint_pkg-android-tools-devel
-
 maint_pkg-erlang-devel
maint_pkg-erlang-devel
-
 maint_pkg-fonts-devel
maint_pkg-fonts-devel
-
 maint_pkg-games-devel
maint_pkg-games-devel
-
 maint_pkg-golang-maintainers
maint_pkg-golang-maintainers
-
 maint_pkg-grass-devel
maint_pkg-grass-devel
-
 maint_pkg-haskell-maintainers
maint_pkg-haskell-maintainers
-
 maint_pkg-java-maintainers
maint_pkg-java-maintainers
-
 maint_pkg-javascript-devel
maint_pkg-javascript-devel
-
 maint_pkg-multimedia-maintainers
maint_pkg-multimedia-maintainers
-
 maint_pkg-perl-maintainers
maint_pkg-perl-maintainers
-
 maint_pkg-php-pear
maint_pkg-php-pear
-
 maint_pkg-openstack
maint_pkg-openstack
-
 maint_pkg-r
maint_pkg-r
-
 maint_pkg-ruby-extras-maintainers
maint_pkg-ruby-extras-maintainers
-
 maint_pkg-rust-maintainers
maint_pkg-rust-maintainers
-
 maint_reproducible-builds
maint_reproducible-builds

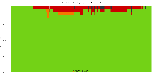
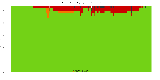
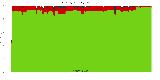
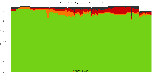
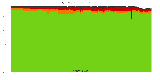
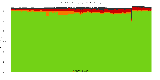
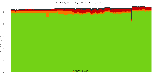
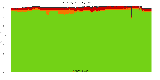
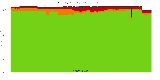
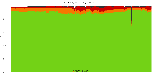
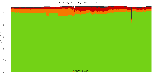
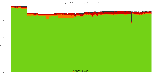
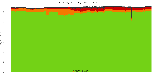
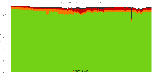
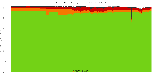
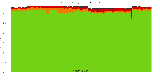
The package set maint_debian-med in
trixie/arm64 consists of 901 packages:
 17 (1.9%) packages
failed to build reproducibly:
metastudent-data
dicom3tools
libpll
hmmer
mapsembler2
ncbi-igblast
parallel
mrtrix3
pydicom
dipy
gdcm
salmon
treeview
librostlab
mmlib
igraph
emboss
17 (1.9%) packages
failed to build reproducibly:
metastudent-data
dicom3tools
libpll
hmmer
mapsembler2
ncbi-igblast
parallel
mrtrix3
pydicom
dipy
gdcm
salmon
treeview
librostlab
mmlib
igraph
emboss
 8 (0.9%) packages
failed to build from source:
picard-tools
pynn
qrisk2
pigx-rnaseq
unifrac-tools
rapmap
q2cliP
python-mne
8 (0.9%) packages
failed to build from source:
picard-tools
pynn
qrisk2
pigx-rnaseq
unifrac-tools
rapmap
q2cliP
python-mne



 37 (4.1%) packages
are either in depwait state, blacklisted, not for us, or cannot be downloaded:
bio-eagle
iqtree
wtdbg2
subarch-select
pftools
mecat2
megahit
fis-gtm
soapdenovo
plink1.9
lambda-align2
bolt-lmm
sortmerna
libstatgen
q2-phylogeny
spades
soapaligner
theseus
probabel
elastix
soapdenovo2
insighttoolkit5
metabat
khmer
obitools
bmtk
xenium
plastimatch
unicycler
minimac4
psortb
sight
libatomic-queue
shovill
libmmmulti
camitk
gubbins
37 (4.1%) packages
are either in depwait state, blacklisted, not for us, or cannot be downloaded:
bio-eagle
iqtree
wtdbg2
subarch-select
pftools
mecat2
megahit
fis-gtm
soapdenovo
plink1.9
lambda-align2
bolt-lmm
sortmerna
libstatgen
q2-phylogeny
spades
soapaligner
theseus
probabel
elastix
soapdenovo2
insighttoolkit5
metabat
khmer
obitools
bmtk
xenium
plastimatch
unicycler
minimac4
psortb
sight
libatomic-queue
shovill
libmmmulti
camitk
gubbins
 839 (93.1%) packages
successfully build reproducibly:
abacas
abpoa
abyss
adapterremoval
adun.app
aegean
aeonbits-owner
aevol
aghermann
alien-hunter
alter-sequence-alignment
altree
amap-align
amide
andi
any2fasta
aragorn
arden
argh
argtable2
ariba
artemis
artfastqgenerator
art-nextgen-simulation-tools
assembly-stats
assemblytics
ataqv
atropos
augur
augustus
autodocksuite
autodock-vina
axe-demultiplexer
baitfisher
bali-phy
bambamc
bamclipper
bamtools
bandage
barrnap
bart
bart-view
bbhash
bbmap
bcalm
bcftools
beads
beagle
beast2-mcmc
beast-mcmc
bedops
bedtools
berkeley-express
bifrost
bioawk
biobambam2
biojava4-live
biojava6-live
biojava-live
biomaj3
biomaj3-cli
biomaj3-core
biomaj3-daemon
biomaj3-download
biomaj3-process
biomaj3-user
biomaj3-zipkin
bioperl
bioperl-run
bio-rainbow
biosig
biosquid
biosyntax
bio-tradis
bio-vcf
bitseq
bowtie
bowtie2
boxshade
bppphyview
bppsuite
brian
brig
btllib
busco
bustools
bwa
camp
capsule-maven-nextflow
capsule-nextflow
cassiopee
castxml
cat-bat
cct
cdbfasta
cd-hit
centrifuge
cgview
changeo
charls
chromhmm
chromimpute
ciftilib
cif-tools
circos
circos-tools
civetweb
clearcut
clonalframe
clonalframeml
clonalorigin
clustalo
clustalw
clustalx
cmtk
cnvkit
codonw
coils
concavity
concurrentqueue
conda-package-handling
conda-package-streaming
consensuscore
conservation-code
covtobed
crac
ctdconverter
ctdopts
ctffind
ctn
ctsim
cutesv
cwlformat
cwltest
cwltool
cycle
cyvcf2
daligner
damapper
dascrubber
dawg
dazzdb
dcm2niix
dcmtk
debian-med
delly
dextractor
dialign
dialign-t
diamond-aligner
dicomscope
discosnp
dnaclust
dnapi
dnarrange
drmaa
drop-seq
dssp
dwgsim
ea-utils
ecopcr
edfbrowser
edflib
edtsurf
eegdev
eigensoft
elph
embassy-domainatrix
embassy-domalign
embassy-domsearch
emboss-explorer
e-mem
emmax
emperor
epcr
estscan
examl
exonerate
fast5
fasta3
fastani
fastaq
fastdnaml
fastlink
fastml
fastp
fastqc
fastq-pair
fastqtl
fasttree
fermi-lite
ffindex
figtree
filtlong
fitgcp
flash
flexbar
flye
freecontact
fsa
fsm-lite
g2
galileo
garli
gasic
gatb-core
gatk-bwamem
gatk-fermilite
gdpc
gemma
genometester
genomethreader
genometools
gentle
getdata
gfapy
gff2aplot
gff2ps
gffread
ggd-utils
ghmm
gifticlib
gjh-asl-json
glam2
gmap
gnumed-client
gnumed-server
grabix
graphlan
grinder
gsort
gwama
gwyddion
harmonypy
harvest-tools
hdmf
hhsuite
hinge
hisat2
hmmer2
hnswlib
hopscotch-map
htscodecs
htseq
htsjdk
htslib
hts-nim-tools
hunspell-en-med
hyphy
icb-utils
idba
igor
igv
illustrate
imagej
indelible
infernal
insilicoseq
ipig
ismrmrd#
iva
ivar
jaligner
jam-lib
jebl2
jellyfish
jellyfish1
jheatchart
jmodeltest
kalign
kallisto
kaptive
kineticstools
king
king-probe
kissplice
kleborate
klustakwik
kma
kmc
kmer
kmerresistance
kraken
kraken2
lagan
lamarc
lamassemble
lambda-align
last-align
lastz
lefse
libace-perl
libamplsolver
libargparse
libargs
libatomicbitvector
libbigwig
libbio-alignio-stockholm-perl
libbio-cluster-perl
libbio-coordinate-perl
libbio-db-biofetch-perl
libbio-db-embl-perl
libbio-db-hts-perl
libbio-db-ncbihelper-perl
libbio-db-seqfeature-perl
libbio-featureio-perl
libbio-graphics-perl
libbio-mage-perl
libbio-mage-utils-perl
libbioparser-dev
libbiosoup-dev
libbio-tools-run-remoteblast-perl
libbio-variation-perl
libbitarray
libbpp-core
libbpp-phyl
libbpp-phyl-omics
libbpp-popgen
libbpp-qt
libbpp-raa
libbpp-seq
libbpp-seq-omics
libcereal
libchado-perl
libcifpp
libcolt-free-java
libctapimkt
libdeflate
libdisorder
libdistlib-java
libdivsufsort
libedlib
libfastahack
libfreecontact-perl
libgclib
libgdf
libgenome
libgenome-model-tools-music-perl
libgenome-perl
libgff
libgkarrays
libgoby-java
libgo-perl
libgtextutils
libgzstream
libhac-java
libhat-trie
libhmsbeagle
libhpptools
libics
libips4o
libjloda-java
libjung-free-java
libkmlframework-java
libla4j-java
libleidenalg
liblemon
libmaus2
libmcfp
libmems
libmialm
libminc
libmjson-java
libmmap-allocator
libmurmurhash
libmuscle
libncl
libnewuoa
liboptions-java
libpal-java
libpj-java
libpsortb
libqes
librandom123
librcsb-core-wrapper
librdp-taxonomy-tree-java
librg-blast-parser-perl
librg-exception-perl
librg-utils-perl
librostlab-blast
libsbml
libsecrecy
libseqlib
libshrinkwrap
libsis-base-java
libsis-jhdf5-java
libslow5lib
libsmithwaterman
libsort-key-top-perl
libssw
libstreamvbyte
libtecla
libtfbs-perl
libthread-pool
libvbz-hdf-plugin
libvcflib
libvistaio
libwfa2
libxdf
libzeep
libzerg
lighter
loki
ltrsift
lucy
lumpy-sv
maffilter
mafft
malt
mapdamage
maq
maqview
mash
maude
mauve-aligner
mcaller
mcl
mcl14
medicalterms
megadepth
megan-ce
melting
mencal
metaeuk
metaphlan
metaphlan2-data
metastudent
metastudent-data-2
mhap
microbegps
microbiomeutil
milib
minc-tools
mindthegap
minia
miniasm
minimap
minimap2
mipe
mira
mirtop
mmseqs2
mosdepth
mothur
mptp
mrbayes
mrc
mricron
multiqc
mummer
muscle
muscle3
mustang
nanofilt
nanolyse
nanook
nanopolish
nanosv
ncbi-acc-download
ncbi-entrez-direct
ncbi-tools6
ncbi-vdb
neo
neobio
ngmlr
nibabel
nifticlib
nim-hts
nim-kexpr
nim-lapper
nipy
njplot
norsnet
norsp
ntcard
nthash
nutsqlite
odil
odin
ont-fast5-api
opencfu
openslide
openslide-python
optimir
orthanc
orthanc-dicomweb
orthanc-gdcm
orthanc-imagej
orthanc-mysql
orthanc-neuro
orthanc-postgresql
orthanc-python
orthanc-webviewer
orthanc-wsi
oscar
pairtools
pal2nal
paleomix
paml
papyrus
paraclu
parafly
parallel-fastq-dump
parasail
parsnp
paryfor
patman
patsy
pbbam
pbcopper
pbdagcon
pbseqlib
pbsim
pcalendar
pdb2pqr
peptidebuilder
perlprimer
perm
phast
phipack
phybin
phylip
phylonium
phyml
physamp
phyutility
phyx
picopore
piler
pilercr
pilon
pinfish
pique
pirs
pixelmed
pixelmed-codec
pizzly
placnet
plasmidid
plasmidomics
plasmidseeker
plast
plink
plink2
plip
poa
populations
porechop
poretools
praat
prank
predictnls
presto
prime-phylo
primer3
prinseq-lite
proalign
probalign
probcons
proda
prodigal
profbval
profisis
profnet
proftmb
progressivemauve
prokka
propka
proteinortho
prottest
pscan-chip
pscan-tfbs
psignifit
psychopy
pullseq
pvrg-jpeg
pybel
pybigwig
pycoqc
pycorrfit
pyensembl
pyfastx
pynwb
pyode
pyqi
pyranges
pyscanfcs
pysurfer
python-alignlib
python-bcbio-gff
python-bel-resources
python-bids-validator
python-bioblend
python-bioframe
python-biom-format
python-biopython
python-biotools
python-bx
python-cgecore
python-cgelib
python-cigar
python-cobra
python-csb
python-cutadapt
python-cykhash
python-datacache
python-deeptools
python-deeptoolsintervals
python-dendropy
python-dicompylercore
python-dnaio
python-ete3
python-etelemetry
python-fitbit
python-freecontact
python-geneimpacts
python-gffutils
python-gtfparse
python-hl7
python-hmmlearn
python-iow
python-leidenalg
python-nanoget
python-nanomath
python-pairix
python-pangolearn
python-parasail
python-pauvre
python-pbcommand
python-pbcore
python-prefixed
python-py2bit
python-pyani
python-pybedtools
python-pycosat
python-pyfaidx
python-pyflow
python-pymummer
python-pysam
python-pyspoa
python-pyvcf
python-questplus
python-ruffus
python-scitrack
python-screed
python-skbio
python-sqt
python-tinyalign
python-treetime
python-trx-python
python-wdlparse
pyxid
pyxnat
q2-alignment
q2-cutadapt
q2-diversity-lib
q2-emperor
q2-feature-table
q2-fragment-insertion
q2-metadata
q2-quality-filter
q2-sample-classifier
q2-taxa
q2templates
q2-types
qcat
qcumber
qiime
qsopt-ex
qtltools
quicktree
quitcount
quorum
racon
radiant
ragout
rambo-k
rampler
rate4site
raxml
ray
rdp-alignment
rdp-classifier
rdp-readseq
readerwriterqueue
readseq
reapr
recan
relacy
relion
repeatmasker-recon
resfinder
resfinder-db
rnahybrid
rna-star
roary
rockhopper
roguenarok
rsem
rtax
ruby-rgfa
runcircos-gui
saint
salmid
sambamba
samblaster
samtools
samtools-legacy
savvy
sbmltoolbox
scoary
scrappie
scrm
scythe
seaview
seer
segemehl
seirsplus
seqan2
seqan3
seqan-needle
seqan-raptor
seqkit
seqmagick
seqprep
seqsero
seqtk
seqtools
seriation
setcover
sga
shapeit4
shasta
sibelia
sibsim4
sigma-align
sigviewer
sim4
simde
simka
simrisc
ska
skesa
skewer
smalt
snakemake
snap
snap-aligner
sniffles
snpeff
snpomatic
snpsift
snp-sites
soapsnp
socket++
sorted-nearest
spaced
spaln
spdlog
spoa
spread-phy
squizz
sra-sdk
srf
srst2
ssake
sspace
ssshtest
stacks
staden
staden-io-lib
stringtie
subread
suitename
sumaclust
sumalibs
sumatra
surankco
surpyvor
survivor
svim
swarm-cluster
sweed
swissknife
tantan
tao-config
tao-json
t-coffee
terraphast
thesias
tiddit
tigr-glimmer
tm-align
tnseq-transit
toil
tombo
toml11
tophat-recondition
toppred
tortoize
trace2dbest
tracetuner
transdecoder
transrate-tools
transtermhp
tree-puzzle
treeviewx
trim-galore
trimmomatic
trinculo
trinityrnaseq
tvc
uc-echo
ugene
umis
uncalled
unifrac
unikmer
varna
vcfanno
vcftools
velvet
velvetoptimiser
veryfasttree
virulencefinder
vmatch
volpack
vsearch
vt
wham-align
wise
xdffileio
xmedcon
xpore
xxsds-dynamic
yaggo
yaha
yanagiba
yanosim
839 (93.1%) packages
successfully build reproducibly:
abacas
abpoa
abyss
adapterremoval
adun.app
aegean
aeonbits-owner
aevol
aghermann
alien-hunter
alter-sequence-alignment
altree
amap-align
amide
andi
any2fasta
aragorn
arden
argh
argtable2
ariba
artemis
artfastqgenerator
art-nextgen-simulation-tools
assembly-stats
assemblytics
ataqv
atropos
augur
augustus
autodocksuite
autodock-vina
axe-demultiplexer
baitfisher
bali-phy
bambamc
bamclipper
bamtools
bandage
barrnap
bart
bart-view
bbhash
bbmap
bcalm
bcftools
beads
beagle
beast2-mcmc
beast-mcmc
bedops
bedtools
berkeley-express
bifrost
bioawk
biobambam2
biojava4-live
biojava6-live
biojava-live
biomaj3
biomaj3-cli
biomaj3-core
biomaj3-daemon
biomaj3-download
biomaj3-process
biomaj3-user
biomaj3-zipkin
bioperl
bioperl-run
bio-rainbow
biosig
biosquid
biosyntax
bio-tradis
bio-vcf
bitseq
bowtie
bowtie2
boxshade
bppphyview
bppsuite
brian
brig
btllib
busco
bustools
bwa
camp
capsule-maven-nextflow
capsule-nextflow
cassiopee
castxml
cat-bat
cct
cdbfasta
cd-hit
centrifuge
cgview
changeo
charls
chromhmm
chromimpute
ciftilib
cif-tools
circos
circos-tools
civetweb
clearcut
clonalframe
clonalframeml
clonalorigin
clustalo
clustalw
clustalx
cmtk
cnvkit
codonw
coils
concavity
concurrentqueue
conda-package-handling
conda-package-streaming
consensuscore
conservation-code
covtobed
crac
ctdconverter
ctdopts
ctffind
ctn
ctsim
cutesv
cwlformat
cwltest
cwltool
cycle
cyvcf2
daligner
damapper
dascrubber
dawg
dazzdb
dcm2niix
dcmtk
debian-med
delly
dextractor
dialign
dialign-t
diamond-aligner
dicomscope
discosnp
dnaclust
dnapi
dnarrange
drmaa
drop-seq
dssp
dwgsim
ea-utils
ecopcr
edfbrowser
edflib
edtsurf
eegdev
eigensoft
elph
embassy-domainatrix
embassy-domalign
embassy-domsearch
emboss-explorer
e-mem
emmax
emperor
epcr
estscan
examl
exonerate
fast5
fasta3
fastani
fastaq
fastdnaml
fastlink
fastml
fastp
fastqc
fastq-pair
fastqtl
fasttree
fermi-lite
ffindex
figtree
filtlong
fitgcp
flash
flexbar
flye
freecontact
fsa
fsm-lite
g2
galileo
garli
gasic
gatb-core
gatk-bwamem
gatk-fermilite
gdpc
gemma
genometester
genomethreader
genometools
gentle
getdata
gfapy
gff2aplot
gff2ps
gffread
ggd-utils
ghmm
gifticlib
gjh-asl-json
glam2
gmap
gnumed-client
gnumed-server
grabix
graphlan
grinder
gsort
gwama
gwyddion
harmonypy
harvest-tools
hdmf
hhsuite
hinge
hisat2
hmmer2
hnswlib
hopscotch-map
htscodecs
htseq
htsjdk
htslib
hts-nim-tools
hunspell-en-med
hyphy
icb-utils
idba
igor
igv
illustrate
imagej
indelible
infernal
insilicoseq
ipig
ismrmrd#
iva
ivar
jaligner
jam-lib
jebl2
jellyfish
jellyfish1
jheatchart
jmodeltest
kalign
kallisto
kaptive
kineticstools
king
king-probe
kissplice
kleborate
klustakwik
kma
kmc
kmer
kmerresistance
kraken
kraken2
lagan
lamarc
lamassemble
lambda-align
last-align
lastz
lefse
libace-perl
libamplsolver
libargparse
libargs
libatomicbitvector
libbigwig
libbio-alignio-stockholm-perl
libbio-cluster-perl
libbio-coordinate-perl
libbio-db-biofetch-perl
libbio-db-embl-perl
libbio-db-hts-perl
libbio-db-ncbihelper-perl
libbio-db-seqfeature-perl
libbio-featureio-perl
libbio-graphics-perl
libbio-mage-perl
libbio-mage-utils-perl
libbioparser-dev
libbiosoup-dev
libbio-tools-run-remoteblast-perl
libbio-variation-perl
libbitarray
libbpp-core
libbpp-phyl
libbpp-phyl-omics
libbpp-popgen
libbpp-qt
libbpp-raa
libbpp-seq
libbpp-seq-omics
libcereal
libchado-perl
libcifpp
libcolt-free-java
libctapimkt
libdeflate
libdisorder
libdistlib-java
libdivsufsort
libedlib
libfastahack
libfreecontact-perl
libgclib
libgdf
libgenome
libgenome-model-tools-music-perl
libgenome-perl
libgff
libgkarrays
libgoby-java
libgo-perl
libgtextutils
libgzstream
libhac-java
libhat-trie
libhmsbeagle
libhpptools
libics
libips4o
libjloda-java
libjung-free-java
libkmlframework-java
libla4j-java
libleidenalg
liblemon
libmaus2
libmcfp
libmems
libmialm
libminc
libmjson-java
libmmap-allocator
libmurmurhash
libmuscle
libncl
libnewuoa
liboptions-java
libpal-java
libpj-java
libpsortb
libqes
librandom123
librcsb-core-wrapper
librdp-taxonomy-tree-java
librg-blast-parser-perl
librg-exception-perl
librg-utils-perl
librostlab-blast
libsbml
libsecrecy
libseqlib
libshrinkwrap
libsis-base-java
libsis-jhdf5-java
libslow5lib
libsmithwaterman
libsort-key-top-perl
libssw
libstreamvbyte
libtecla
libtfbs-perl
libthread-pool
libvbz-hdf-plugin
libvcflib
libvistaio
libwfa2
libxdf
libzeep
libzerg
lighter
loki
ltrsift
lucy
lumpy-sv
maffilter
mafft
malt
mapdamage
maq
maqview
mash
maude
mauve-aligner
mcaller
mcl
mcl14
medicalterms
megadepth
megan-ce
melting
mencal
metaeuk
metaphlan
metaphlan2-data
metastudent
metastudent-data-2
mhap
microbegps
microbiomeutil
milib
minc-tools
mindthegap
minia
miniasm
minimap
minimap2
mipe
mira
mirtop
mmseqs2
mosdepth
mothur
mptp
mrbayes
mrc
mricron
multiqc
mummer
muscle
muscle3
mustang
nanofilt
nanolyse
nanook
nanopolish
nanosv
ncbi-acc-download
ncbi-entrez-direct
ncbi-tools6
ncbi-vdb
neo
neobio
ngmlr
nibabel
nifticlib
nim-hts
nim-kexpr
nim-lapper
nipy
njplot
norsnet
norsp
ntcard
nthash
nutsqlite
odil
odin
ont-fast5-api
opencfu
openslide
openslide-python
optimir
orthanc
orthanc-dicomweb
orthanc-gdcm
orthanc-imagej
orthanc-mysql
orthanc-neuro
orthanc-postgresql
orthanc-python
orthanc-webviewer
orthanc-wsi
oscar
pairtools
pal2nal
paleomix
paml
papyrus
paraclu
parafly
parallel-fastq-dump
parasail
parsnp
paryfor
patman
patsy
pbbam
pbcopper
pbdagcon
pbseqlib
pbsim
pcalendar
pdb2pqr
peptidebuilder
perlprimer
perm
phast
phipack
phybin
phylip
phylonium
phyml
physamp
phyutility
phyx
picopore
piler
pilercr
pilon
pinfish
pique
pirs
pixelmed
pixelmed-codec
pizzly
placnet
plasmidid
plasmidomics
plasmidseeker
plast
plink
plink2
plip
poa
populations
porechop
poretools
praat
prank
predictnls
presto
prime-phylo
primer3
prinseq-lite
proalign
probalign
probcons
proda
prodigal
profbval
profisis
profnet
proftmb
progressivemauve
prokka
propka
proteinortho
prottest
pscan-chip
pscan-tfbs
psignifit
psychopy
pullseq
pvrg-jpeg
pybel
pybigwig
pycoqc
pycorrfit
pyensembl
pyfastx
pynwb
pyode
pyqi
pyranges
pyscanfcs
pysurfer
python-alignlib
python-bcbio-gff
python-bel-resources
python-bids-validator
python-bioblend
python-bioframe
python-biom-format
python-biopython
python-biotools
python-bx
python-cgecore
python-cgelib
python-cigar
python-cobra
python-csb
python-cutadapt
python-cykhash
python-datacache
python-deeptools
python-deeptoolsintervals
python-dendropy
python-dicompylercore
python-dnaio
python-ete3
python-etelemetry
python-fitbit
python-freecontact
python-geneimpacts
python-gffutils
python-gtfparse
python-hl7
python-hmmlearn
python-iow
python-leidenalg
python-nanoget
python-nanomath
python-pairix
python-pangolearn
python-parasail
python-pauvre
python-pbcommand
python-pbcore
python-prefixed
python-py2bit
python-pyani
python-pybedtools
python-pycosat
python-pyfaidx
python-pyflow
python-pymummer
python-pysam
python-pyspoa
python-pyvcf
python-questplus
python-ruffus
python-scitrack
python-screed
python-skbio
python-sqt
python-tinyalign
python-treetime
python-trx-python
python-wdlparse
pyxid
pyxnat
q2-alignment
q2-cutadapt
q2-diversity-lib
q2-emperor
q2-feature-table
q2-fragment-insertion
q2-metadata
q2-quality-filter
q2-sample-classifier
q2-taxa
q2templates
q2-types
qcat
qcumber
qiime
qsopt-ex
qtltools
quicktree
quitcount
quorum
racon
radiant
ragout
rambo-k
rampler
rate4site
raxml
ray
rdp-alignment
rdp-classifier
rdp-readseq
readerwriterqueue
readseq
reapr
recan
relacy
relion
repeatmasker-recon
resfinder
resfinder-db
rnahybrid
rna-star
roary
rockhopper
roguenarok
rsem
rtax
ruby-rgfa
runcircos-gui
saint
salmid
sambamba
samblaster
samtools
samtools-legacy
savvy
sbmltoolbox
scoary
scrappie
scrm
scythe
seaview
seer
segemehl
seirsplus
seqan2
seqan3
seqan-needle
seqan-raptor
seqkit
seqmagick
seqprep
seqsero
seqtk
seqtools
seriation
setcover
sga
shapeit4
shasta
sibelia
sibsim4
sigma-align
sigviewer
sim4
simde
simka
simrisc
ska
skesa
skewer
smalt
snakemake
snap
snap-aligner
sniffles
snpeff
snpomatic
snpsift
snp-sites
soapsnp
socket++
sorted-nearest
spaced
spaln
spdlog
spoa
spread-phy
squizz
sra-sdk
srf
srst2
ssake
sspace
ssshtest
stacks
staden
staden-io-lib
stringtie
subread
suitename
sumaclust
sumalibs
sumatra
surankco
surpyvor
survivor
svim
swarm-cluster
sweed
swissknife
tantan
tao-config
tao-json
t-coffee
terraphast
thesias
tiddit
tigr-glimmer
tm-align
tnseq-transit
toil
tombo
toml11
tophat-recondition
toppred
tortoize
trace2dbest
tracetuner
transdecoder
transrate-tools
transtermhp
tree-puzzle
treeviewx
trim-galore
trimmomatic
trinculo
trinityrnaseq
tvc
uc-echo
ugene
umis
uncalled
unifrac
unikmer
varna
vcfanno
vcftools
velvet
velvetoptimiser
veryfasttree
virulencefinder
vmatch
volpack
vsearch
vt
wham-align
wise
xdffileio
xmedcon
xpore
xxsds-dynamic
yaggo
yaha
yanagiba
yanosim
A package name displayed with a bold
font is an indication that this package has a note. Visited
packages are linked in green, those which have not been visited are
linked in blue.
A # sign after the name of a package
indicates that a bug is filed against it. Likewise, a
+ sign indicates there is a
patch available, a P means a
pending bug while # indicates a
closed bug. In cases of several bugs, the symbol is repeated.